Nanobody CDR3 structure prediction
Four years after the protein folding problem was allegedly solved, we still can’t reliably predict how or where antibodies bind to their antigens. A recent report identifies one source of continued difficulty.
Nanobodies are single-domain antibodies used as clinical therapeutics and experimental reagents for tasks such as bulking up protein structures for cryo-EM [1]. Despite their smaller size, they share the same difficulties as traditional antibodies when it comes to structure prediction, namely the inability to reliably predict how and where they bind their targets, with success rates ranging in the 33-50% range [2, 3]. A report published last week [4] identifies a major determinant of whether the binding mode is correctly predicted: the “kink” or bend found in many CDR3 loops that is also found in most human antibodies [5]. The recent structure prediction tool AlphaFold3 correctly docked nanobodies to their antigens nearly 80% of the time when the CDR3 conformation was extended, but barely 25% of the time when it was kinked.
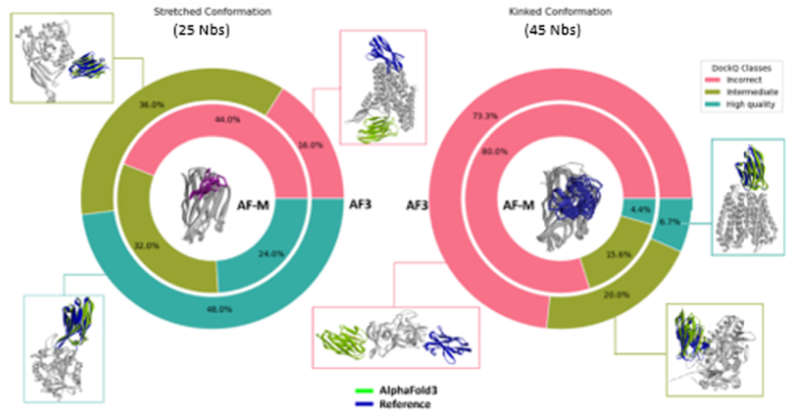 Figure from [4]
Figure from [4]
Fortunately, these models were effective at predicting whether the kink was there or not.
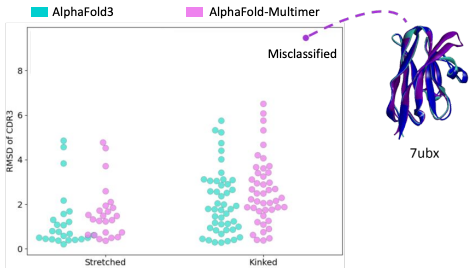 Figure from [4]
Figure from [4]
In any case, whether that observation extends to human paired antibodies, the structures of which are more difficult to predict [6], remains to be seen.
- Wu, Xudong. “Nanobody-assisted cryoEM structural determination for challenging proteins” Trends in Biochemical Sciences 2024
- Abramson, Josh, et al. “Accurate structure prediction of biomolecular interactions with AlphaFold3” Nature 2024
- Hitawala, Fatima N. and Gray, Jeffrey J. “What has AlphaFold3 learned about antibody and nanobody docking, and what remains unsolved?” Biorxiv 2024
- Eshak, Floriane and Goupil-Lamy, Anne. “Advancements in nanobody epitope prediction: A comparative study of AlphaFold2Multimer vs AlphaFold3” JCIM 2025
- Weitzner, Brian D. and Dunbrack, Roland L. and Gray, Jeffrey J. “The origin of CDR H3 structural diversity” Structure 2015
- Greenshields-Watson, et al. “Challenges and compromises: Predicting unbound antibody structures with deep learning” Current Opinion in Structural Biology 2025