Structural adaptors for protein property prediction
It’s always tricky to choose which protein neural network to use for fine-tuning tasks.
Inverse folding neural networks like ProteinMPNN [1] have the advantage of working with explicit protein structures, but don’t leverage the huge sequence databases we have available, whereas the opposite is true for protein language models like ESM [2]. Some methods, such as ESM-IF1 [3], try to bridge the gap by predicting structures from sequence and training from those, but those predicted structures aren’t as informative as experimental structures [4]. In any case, it’s clear that structure-based neural networks are best for zero-shot prediction of things like stability and protein-protein binding affinity prediction, whereas sequence-based neural networks are better at predicting things like catalytic activity and overall contribution to organismal fitness [5].
A recent preprint by Li & Luo [6] discusses a way to combine both types of neural networks when fine-tuning for protein property prediction. Their method, names SPURS, introduces the final embeddings from the structure-based neural network ProteinMPNN into each intermediate layer of ESM2, thereby passing on the advantages of that 3D information to the sequence-based model.
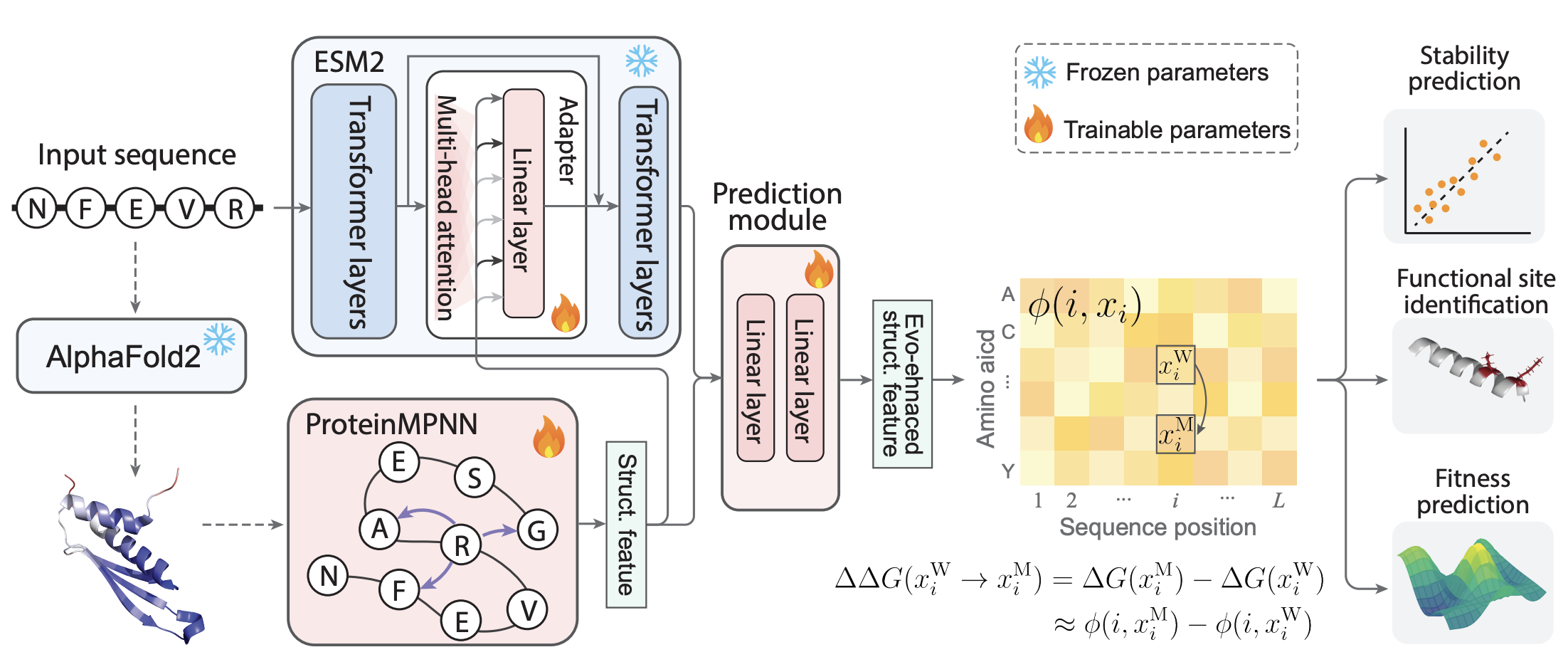 Figure from [6]
Figure from [6]
After fine-tuning this network on the mega-scale dataset by Tsuboyama et al [7], which measured nearly a million stability measurements across several hundred proteins, they end up with a state-of-the-art thermostability predictor. This beats ThermoMPNN, a fine-tuned version of ProteinMPNN trained on the same dataset [8], directly showing how adding the language model into the mix contributed to performance.
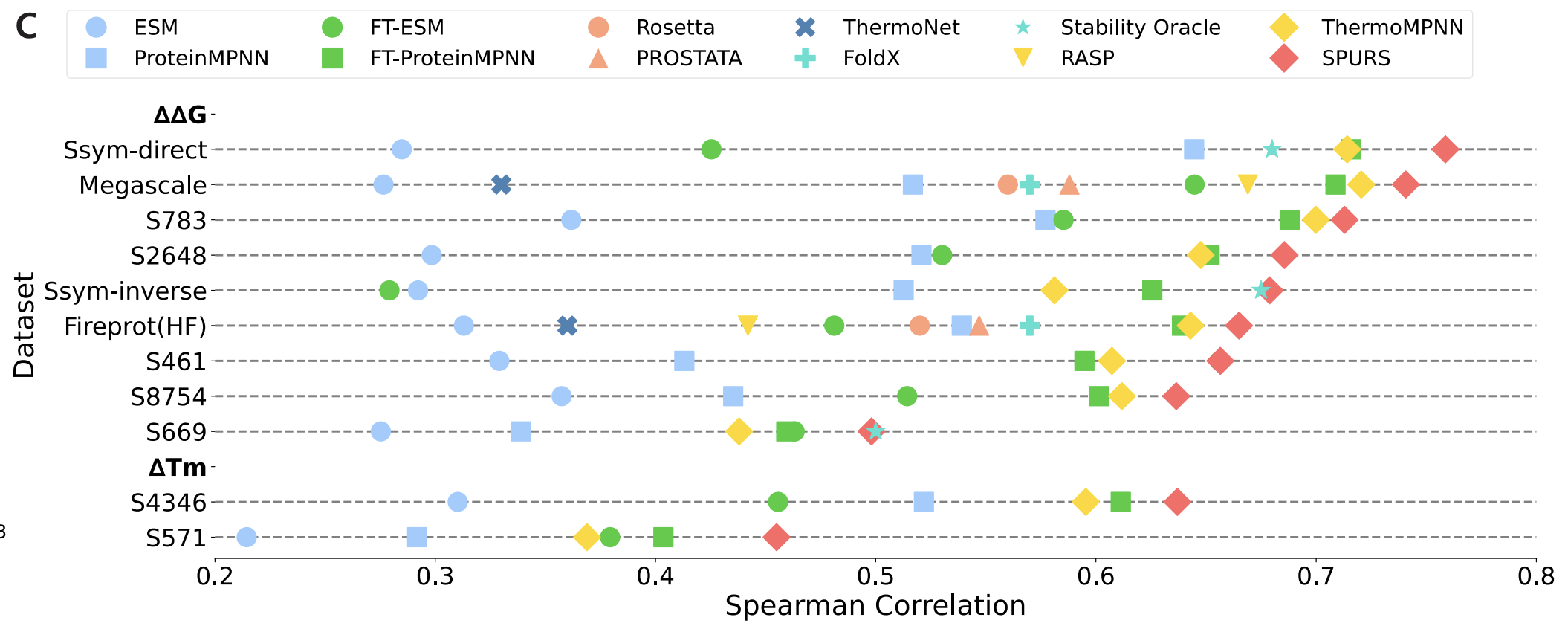 Figure from [6]
Figure from [6]
This isn’t the first time structural adaptors for language models have been discussed; Ruffolo et al [9] discussed the idea for generative modeling using the ProGen language model late last year. But this is (to my knowledge) the first time I’ve seen it for this kind of property prediction. It’s also worth noting that this scheme only works when large-scale data is available, as it adds thousands of parameters that would, in a normal experimental setup, be underdetermined without some kind of regularization.
- Dauparas et al. “Robust deep learning-based protein sequence design using ProteinMPNN” Science 2022
- Lin et al. “Evolutionary-scale prediction of atomic-level protein structure with a language model” Science 2023
- Hsu et al. “Learning inverse folding from millions of predicted structures” ICML 2022
- Su et al. “SaProt: Protein language modeling with structure-aware vocabulary” ICLR 2024
- Notin et al. “ProteinGym: Large-scale benchmarks for protein design and fitness prediction” biorxiv 2023
- Li & Luo. “Rewiring protein sequence and structure generative models to enhance protein stability prediction” biorxiv 2025
- Tsuboyama et al. “Mega-scale experimental analysis of protein folding stability in biology and protein design” Nature 2023
- Dieckhaus et al. “Transfer learning to leverage larger datasets for improved prediction of protein stability changes” PNAS 2024
- Ruffolo et al. “Adapting protein language models for structure-conditioned design” biorxiv 2024